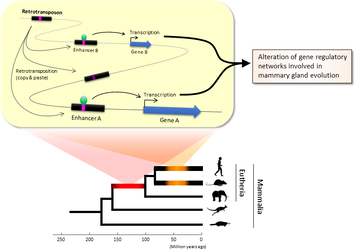
 Figure 1. Retrotransposons have been co-opted to act as enhancers in the mammalian genome. Transposable elements known as retrotransposons duplicate themselves via a copy-and-paste mechanism. Tens of thousands of these sequences were found to contain binding sites for proteins that act as master regulators of mammary gland development. Figure 1. Retrotransposons have been co-opted to act as enhancers in the mammalian genome. Transposable elements known as retrotransposons duplicate themselves via a copy-and-paste mechanism. Tens of thousands of these sequences were found to contain binding sites for proteins that act as master regulators of mammary gland development. Transposable elements (TEs), so-called selfish DNA sequences, are known to be capable of moving around the genome through cut-and-paste or copy-and-paste mechanisms, and our human genome contains approx 4.5 million copies of these TEs. This can not be termed as one obscure event as they account for 30-50% of mammalian DNA. The trnasposable elements have been traditionally considered as genetic freeloaders hitchhiking along in the genome without providing any benefit to the host organism. More recently, however, scientists have begun to uncover cases in which TE sequences have been co-opted by the host to provide a useful function, such as encoding part of a host protein. In a recent study published in the journal Nucleic Acids Research, Professor Hidenori Nishihara from Department of Life Science and Technology, Tokyo Institute of Technology, who has undertaken one of the most comprehensive analyses of TE sequence co-option to date, uncovers tens of thousands of potentially co-opted TE sequences and the findings suggest that the TEs might have played a key role in mammalian evolution. Talking about his research Professor Nishihara says that "I was specifically interested in the potential influence of TE sequences on the evolution of the mammary gland, an organ that is responsible for producing milk and is, as the name suggests, a key distinguishing feature of mammals." To identify potentially co-opted TE sequences, Dr. Nishihara used four proteins—ERα, FoxA1, GATA3, and AP2γ—that bind to DNA to regulate the production of proteins involved in mammary gland development, and located all of the DNA sequences in the genome to which these proteins bind. Surprisingly, 20–30% of all of the binding sites across the genome were located in TEs, with as many as 38,500 TEs containing at least one binding site. The majority of these were in a copy-and-paste type of TE known as a retrotransposon, which duplicates itself, leaving a new copy in a new location. The TE-derived binding site sequences were more conserved across species than expected, indicating that they are being preserved by evolution because they serve some important function. Dr. Nishihara believes that these TE sequences have been co-opted to serve as enhancers, DNA elements that increase the transcription of nearby genes (Fig. 1). By binding to one of the four master regulators of mammary gland development, these enhancers ultimately increase the production of proteins involved in mammary gland development. Dr. Nishihara then investigated when in mammalian evolution these TE sequences were acquired and found two distinct phases of acquisition: roughly 60–70% were acquired in the ancestor of all placental mammals (Eutheria), while 10–20% could be traced back to the ancestor of New World monkeys (Simiiformes) (Fig. 2, left). In addition, there appeared to be another wave of acquisition of ERα binding sites in the ancestor of mice and rats (Muridae) (Fig. 2, right). Thus, by providing a vast number of potential regulatory element binding sites throughout the genome, TEs may have had a substantial impact on the emergence of the mammary gland and its evolution within mammals. 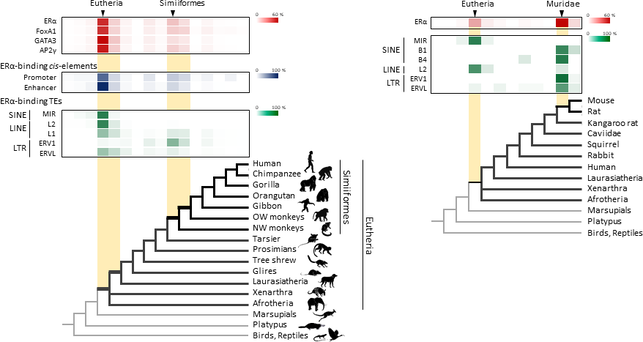 Figure 2. Transposable element-derived binding sites were acquired during distinct phases in mammalian evolution. Left: Among the TE-derived binding sites identified, 60–70% were acquired in the ancestor of placental mammals (Eutheria), while 10–20% were acquired in the ancestor of New World monkeys (Simiiformes). Right: Many ERα binding sites were also acquired in the ancestor of mice and rats (Muridae). Dr. Nishihara's study sheds light on the deep involvement of TEs in the evolution of mammary gland regulatory elements. However, it remains unclear how common this mode of TE-mediated regulatory network evolution is. Dr. Nishihara, at least, believes that the mammary gland is not unique in this respect. He notes that, "in addition to mammary glands, mammals share many features, such as the neocortex, closed secondary palate, and hair. I expect future research to uncover many additional kinds of TEs that have been similarly involved in the evolution of these features in mammals." References
0 Comments
One of the oldest desire of man kind is to increase its life expectancy and if possible be immortal. In pursuit of this dream countless have spent their lives searching for elixir of immortality to fountains of youth, often leading to pain and animosity between fellow humans. Japan, a country owing to its cultural, behavioral and numerous genetic factors has been blessed with many centenarians and currently has the greatest number of known centenarians of any nation with 67,824 according to their 2017 census, along with the highest proportion of centenarians at 34.85 per 100,000 people. Thus Japan becomes the primal choice for conducting studies reflecting on the secrets of longer life and it can be performed with accuracy and a larger statistical sample size compared to other nations. In a latest set of findings research teams of scientists from the RIKEN Center for Integrative Medical Science (IMS) and Keio University School of Medicine in Japan have shown us that all the while we have been looking in the wrong direction and the solution and clue to long life was within our body's own defense / the immune system. Supercentenarians -- meaning people over the age of 110 --as their study interest the teams have discovered an interesting finding that the supercentenarians proved to be an unique group of people having a higher count of specific immune cells cytotoxic CD4+ T-cells, when they compared their cell count with a group of supercentenarians and younger controls. They acquired a total of 41,208 cells from seven supercentenarians (an average of 5,887 per subject) and 19,994 cells for controls (an average of 3,999 per subject) from five controls aged in their fifties to eighties. The study revealed two interesting findings:
Kosuke Hashimoto of IMS, the first author of the paper, expressed the team's stand as "We were especially interested in studying this group of people, because we consider them to be a good model of healthy aging, and this is important in societies like Japan where aging is proceeding rapidly." IMS Deputy Director Piero Carninci, one of the leaders of the groups, says, "This research shows how single-cell transcription analysis can help us to understand how individuals are more or less susceptible to diseases. CD4-positive cells generally work by generating cytokines, while CD8-positive cells are cytotoxic, and it may be that the combination of these two features allows these individuals to be especially healthy. We believe that this type of cells, which are relatively uncommon in most individuals, even young, are useful for fighting against established tumors, and could be important for immunosurveillance. This is exciting as it has given us new insights into how people who live very long lives are able to protect themselves from conditions such as infections and cancer." Their research, is published in journal of Proceedings of the National Academy of Sciences (PNAS), and the study was performed by a collaboration including scientists from the RIKEN Center for Integrative Medical Sciences and Keio University School of Medicine. References
|
AuthorHello! My name is Arunabha Banerjee, and I am the mind behind Biologiks. Leaning new things and teaching biology are my hobbies and passion, it is a continuous journey, and I welcome you all to join with me Archives
June 2024
Categories
All
|


 RSS Feed
RSS Feed



