|
Many congratulations Ms. Jyotishree Karmakar, student of Biologiks for securing 1st Class in Botany Hons, Calcutta University. Biologiks is proud of your achievement and wishes you all the best for your future
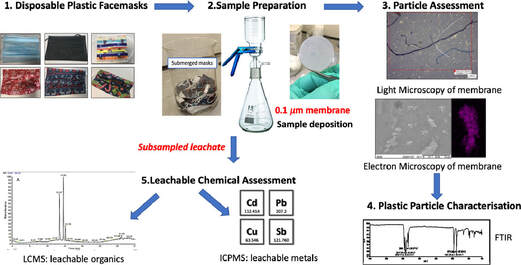 Swansea University scientists have observed leaching of dangerous chemical pollutants from disposable plastic face masks (DPFs)when submerged in water. The research reveals high levels of pollutants, including lead, antimony, and copper, within the silicon-based and plastic fibres of common disposable face masks. Since the outbreak of CoVID-19 use of single use masks along with their associated wastes have risen exponentially as use of mask is the only effective and available resort for everyone, however their usage and discarding have left a deep impact on the environment, and have been documented as a new cause of pollution. The study aimed to explore this direct link – with investigations to identify the level of toxic substances present, and focuses on the emission of pollutants from 7 DPF brands that were submerged in water to simulate environmental conditions if these DPFs were littered. The DPF leachates in water were filtered by inorganic membranes, and both particle-deposited organic membranes and the filtrates were characterized using techniques such as FTIR, SEM-EDX, Light Microscopy, ICP-MS and LC-MS. Micro and nano scale polymeric fibres, particles, siliceous fragments and leachable inorganic and organic chemicals were observed from all of the tested DPFs. Traces concerning heavy metals (i.e. lead up to 6.79 µg/L) were detected in association with silicon containing fragments. ICP-MS also confirmed the presence of other leachable metals like cadmium (up to 1.92 µg/L), antimony (up to 393 µg/L) and copper (up to 4.17 µg/L). LC-MS analysis identified polar leachable organic species related to plastic additives and contaminants; polyamide-66 monomer and oligomers (nylon-66 synthesis), surfactant molecules, dye-like molecules and polyethylene glycol were all tentatively identified in the leachate. The findings reveal significant levels of pollutants in all the masks tested – with micro/nano particles and heavy metals released into the water during all tests. Researchers conclude this will have a substantial environmental impact and, in addition, raise the question of the potential damage to public health – warning that repeated exposure could be hazardous as the substances found have known links to cell death, genotoxicity and cancer formation. Project lead Dr Sarper Sarp of Swansea University College of Engineering said: “All of us need to keep wearing masks as they are essential in ending the pandemic. But we also urgently need more research and regulation on mask production, so we can reduce any risks to the environment and human health”. Dr. Sarper continues: “ improper and unregulated disposal of these DPFs is a plastic pollution problem we are already facing and will only continue to intensify. There is a concerning amount of evidence that suggests that DPFs waste can potentially have a substantial environmental impact by releasing pollutants simply by exposing them to water. Many of the toxic pollutants found in our research have bio-accumulative properties when released into the environment and our findings show that DPFs could be one of the main sources of these environmental contaminants during and after the Covid-19 pandemic. It is, therefore, imperative that stricter regulations need to be enforced during manufacturing and disposal/recycling of DPFs to minimise the environmental impact”. The study also warrants a full investigation is necessary to determine the quantities and potential impacts of these particles leaching into the environment, and the levels being inhaled by users during normal breathing. This is a significant concern, especially for healthcare professionals, key workers, and children who are required to wear masks for large proportions of the working. The group have published their findings in the journal of water research. P.S: Content edited for style and length
Story source: G.L. Sullivan, J. Delgado-Gallardo, T.M. Watson, S. Sarp. An investigation into the leaching of micro and nano particles and chemical pollutants from disposable face masks - linked to the COVID-19 pandemic. Water Research, 2021; 196: 117033 DOI: 10.1016/j.watres.2021.117033 Author: Nitin Bayal National Centre for Cell Science, Pune My presumptions about leprosy were incorrect! Books and documentaries have spoken plenty in silence but still incomplete. In the middle ages, it was one of the most feared diseases and was once considered as “Living Death”. There was a time when prevailing religious concepts kept our attention away from finding the root cause of leprosy transmission and religious institutions influenced the health and disease affairs. Illness due to leprosy was accepted as a curse from God or bad luck, just as many different phenomena that were not linked to cause-and-effect theory credited to the act of god, forcing people to plead and compensate for this unexplained experience. Although after new advances in science and technology came into existence, it became difficult to arguably justify scientific reasoning with superstitions. The dreadful infections in leprosy have much more to say than eradication objectives. Prior to perusing a Ph.D. at National Centre for Cell Science, Pune; I knew very little about leprosy and Mycobacterium leprae, the causative organism of leprosy. Leprosy which causes poor wound healing, nerve damage and if left untreated it results in permanent disfigurement or loss of body parts, has for long been associated with a social stigma. In India, for ages, ayurvedic preparations known as chaulmoogra oil was used to treat leprosy, and with the introduction of western medicine prescription drugs like Dapsone and Rifampicin, the catalog of every possible physical and chemical agent to indiscriminately kill the bad microbes is ever-expanding. It is known to us that a leprosy patient loses the ability to feel the temperature change and pain in the affected area on their body; with the current understanding on the subject, we discovered some mysterious insights on leprosy research that encouraged skin microbiome research in leprosy. We added a brief note on each idea discussed in our research group. I tried to understand the diseased states, current therapeutics, and the problems that challenge scientists in this field. M. leprae slowly gets resistant to new drugs and hence, raises concerns for a more severe form of the disease. In 1982, World Health Organization (WHO) released Multi-Drug Therapy (MDT) guidelines that dictate the planning and execution to eradicate leprosy. There is also the addition of newer drugs into the arsenal along with MDT treatment. Recently, scientists and clinicians speculated India accounts for 66% of leprosy patients worldwide. To challenge leprosy, WHO accelerated a global leprosy strategy for 2016-2020. A team of scientists led by Dr. GP. Talwar at All India Institute of Medical Science (AIIMS); New Delhi developed a vaccine in the 1970s that is finally recommended to launch on 2nd October 2019 after 36 years of testing in the clinical trials. As of now, it is well recognized that leprosy elimination has never been achieved actually in India. Moreover, the frequency of resistant mutants also varies to different drugs, MDT treatment also kills the bacteria that are naturally resistant to drugs on the affected sites. It also affects the normal skin bacterial community. Skin is the primarily affected organ in leprosy. This is where the skin microbiome plays its role to support normal homeostasis. The microbiome consists of an ecosystem of billions or trillions of microorganisms living in and on our bodies including skin. There is growing evidence that skin infections and drug abuse change or disturbs the skin microbiota in various types of skin pathological conditions like acne, eczema, psoriasis, skin allergies, and wounds. We knew that both research scientists and clinicians play a critical role in the successful completion of studies on human subjects through mutual collaborations. While communicating with scientists, dermatologists, and workers of all levels at different NGOs, we finally found our collaborations in 2015 with leprosy clinics at LEPRA and TLM organizations for clinical samples. Then it was followed by regular meetings with patients and leprologists at leprosarium, where patients are kept for their treatment. The situation of patients was horrible because of their constant suffering, thus to understand the relationship between the skin microbiome and M leprae infection, researchers at DBT-National Centre for Cell Science and Bio-Sciences R&D, TCS Innovation Labs, Pune also collaborated to conduct a new study on the skin microbial communities from lesional and non-lesional skin of Indian leprosy patients undergoing multi-drug regimen treatment and healthy individuals covering two different geographical locations viz. Hyderabad and Miraj in India. We obtained ethical and biosafety clearances and collaborated with Dr. Vijayalakshmi Valluri at LEPRA, Hyderabad and, Dr. Rohini Suryavanshi at TLM, Miraj, Maharashtra for clinical samples. We collected skin samples and extracted microbial DNA. We have used DNA sequencing methodologies and analysis tools to define the cutaneous microbiota. Our study has shown that the skin microbiota of healthy persons and leprosy patients have significant differences in its variety and abundance. The reasons also include the role of various external and internal factors comprising skin pH, skin thickness, skin moisture, hygiene, climate, etc. The photograph shown above was taken with consent at Richardson leprosy hospital, Miraj, Maharashtra, and pixelated to maintain the confidentiality of the leprosy patient. Together with Dr. Milind S. Patole as doctoral co-supervisor, Dr. Dhiraj Paul, and Mr. Sunil Nagpal, one of my colleagues at TCS Innovation Labs, we have completed computational analysis on high-throughput sequencing data. We have published the first report on the structure of skin microbiota from India in Springer Nature journals i.e., Nature Scientific Data and Nature Scientific Reports journals. The corresponding authors, Dr. Shekhar C. Mande is Secretary DSIR and Director General, CSIR, New Delhi and Dr. Sharmila S. Mande is Chief Scientist and Head of Bio-Sciences R&D, TCS Research, Pune. They are leading scientists and specialized in Structural Biology and Microbiome research respectively. Our analysis shows differences in the microbiota of affected and unaffected sites on the skin of leprosy patients and similarities among healthy persons irrespective of our sampling sites in India. Microbial diversity probes a distinct depletion of Staphylococcus genera in samples from affected sites of leprosy patients. Major abundant bacteria observed were Pseudomonas, Staphylococcus, Paracoccus, Brevundimonas, Limnobacter, Methylobacterium, Propionibacterium, Corynebacterium, Kocuria, and Streptococcus on the leprosy patient’s skin. It also shows a significant decline in firmicutes-related bacteria in leprosy patients in comparison to healthy individuals. It was a strange coincidence when M leprae attacks and occupy spaces in human bodies by either high exposure to infections or low immunity or both situations. M leprae is very intelligent, being a metabolically challenged bacterium it has managed to survive harsh conditions and still a mystery for the researchers for its cultivation in laboratory conditions. We are also afraid of liberal use of antibiotics and antiseptics that not only results in the extinction of some good bacteria but also kept the fortunate ones under constant threat. The aspect of working with microbiome research can help skin microbiome transplantation science that corresponds to better disease management and diagnosis. A healthy microbiome protects humans from harmful settlers and infections by pathogens or opportunistic microorganisms. Nowadays, many ventures are introducing probiotic products in the skin healthcare system and focusing on implications of skin’s health and appearance. The future of treating skin diseases is hopeful from microbiome research. However, despite these positive developments, measures to improve the healthcare system are also required. Humans often err in their haste. After all the great riches of leprosy are its paradoxes. If we are losing this time that will be another paradox, our luck may also prove contrary. Author Bio: Nitin is a microbiome researcher at DBT-National Institute of Immunology, New Delhi. His interest mainly focus on data analytics in healthcare sector. He is an acclaimed scientific illustrator with a well disposed eye for art in daily lives. Nitin holds a degree of Master of Science in Molecular and Human Genetics and published his Ph.D work on leprosy research. His work is guided by a need to rationalize, make things beautiful, combine science with art and connect ideas.
Area of research: Skin biology, microbiome, sequencing technologies Google Scholar link: https://scholar.google.com/citations?user=79o59fkAAAAJ&hl=en Twitter handle: @BayalNitin Increasing the longevity of indwelling medical implants and preventing Hospital acquired infections11/21/2020 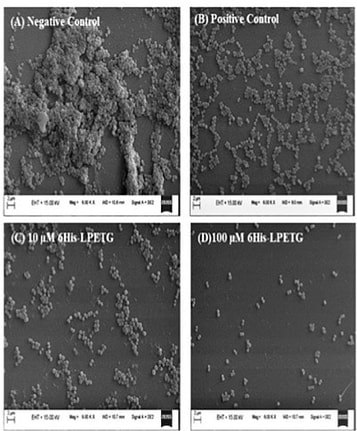 A synthetic peptide (bioconjugate) substrate for cell surface enzyme Sortase A exhibits significant anti-biofilm activity, which can be utilized to make anti-biofilm surfaces for clinical applications and indwelling medical devices. The study conducted by a group of researchers from NIPER Guwahati(National Institute of Pharmaceutical Education and Research, Guwahati), demonstrated that a Sortase-A mediated bioconjugate can inhibit biofilm formation and combat hospital acquired infections, the findings of the study was published in the journal Frontiers in Microbiology. Staphylococcus aureus one of the most notorious pathogens which is frequently associated with nosocomial infections imposing serious risk to immune-compromised patients, because of its ability to colonize at the surface of indwelling medical devices such as catheters, pacemakers, contact lenses, and dentures by biofilm formation. Staphylococcus aureus is known to form multilayered adherent biofilm to the surface of indwelling medical devices including catheters and medical implants, expressing series of toxins which makes them tolerant toward host defense mechanisms and common antibiotics and Biofilm formation on both natural and artificial surfaces is one of the most important virulence mechanisms of many bacterial pathogens, as it guards the bacteria against antibiotic therapy, and thus considered to be the major cause of nosocomial infections especially in post-surgical and immune-compromised patients and is associated with significant mortality in hospitalized patients. Sortase A is one of the most important enzymes present on the cell surface of Gram positive bacteria including S. aureus. Sortase A recognizes the LPXTG motif present at the C-terminus of cell surface proteins and recruits them to the peptidoglycan cell wall building block, lipid II, thus search for molecules that can inhibit Sortase A is one of the promising approaches for the development of innovative strategies to impair bacterial virulence and biofilm formation In the present study, the study group synthesized a novel 6His-LPETG peptide and incorporated it on the cell wall of S. aureus and evaluated the ability of this peptide to inhibit biofilm formation by Gram positive bacteria. Their study exhibits significant anti-biofilm activity of this peptide having an LPETG motif, and it shows that the present mechanism could further be tested against other Gram positive bacteria having Sortase A enzyme. The research team further envisioned that the 6-His epitope along with LPETG motif will allow grafting the surface of the bacteria with epitopes which can be utilized to target the bacteria using 6-His antibodies. In their communication the researchers explained that, "the advantage of our strategy is that it is neither altering any biological process within the bacteria nor inhibiting it. Thus, it is not putting any selective pressure on bacterial population which is one of the major concerns of antibiotic therapy. Our peptide is using bacterial machinery to recruit itself on the cell surface and then hinder the process of biofilm formation". Thus, the strategy can potentially be developed and utilized to make anti-biofilm surfaces for clinical applications. *The study was funded by NIPER seed fund, Government of India* Story source:
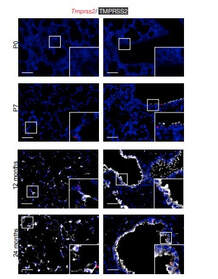
Poonam Kumari, Yutika Nath, Upadhyayula Surayanarayana Murty, Velayutham Ravichandiran and Utpal Mohan. Sortase A Mediated Bioconjugation of Common Epitopes Decreases Biofilm Formation in Staphylococcus aureus. Frontiers in Microbiology. 30 July 2020 | https://doi.org/10.3389/fmicb.2020.01702 P.S. Content edited for style and length Increased expression of a cellular enzyme called TMPRSS2, well known for its role in prostate cancer was observed to play a role in older individuals coming in contact with the virion. The recent findings, published in the Journal of Clinical Investigation, by a group of researchers at Vanderbilt University Medical Center (VUMC) and their colleagues have determined a key factor as to why COVID-19 appears to infect and sicken adults and older people preferentially while seeming to spare younger children. The study observes that children have lower levels of an enzyme/co-receptor that SARS-CoV-2, the RNA virus, needs to invade airway epithelial cells in the lung. The study supports efforts to block the enzyme to potentially treat or prevent COVID-19 in older people. Jennifer Sucre, MD, assistant professor of Pediatrics (Neonatology), who led the research with Jonathan Kropski, MD, assistant professor of Medicine, reports that their study provides a biologic rationale for why particularly infants and very young children seem to be less likely to either get infected or to have severe disease symptoms. The study team infers that there is still so much to learn about SARS-CoV-2. But this much is known: that after a viral particle is inhaled into the lungs, protein “spikes” that stick out like nail studs in a soccer ball attach to ACE2, a receptor on the surfaces of certain lung cells. A cellular enzyme called TMPRSS2 chops up the spike protein, enabling the virus to fuse into the cell membrane and “break into” the cell. Once inside, the virus hijacks the cell’s genetic machinery to make copies of its RNA genome. Dr. Sucre and Dr. Kropski, have collaborated since 2016 and studied lung diseases in premature infants and adults, the epidemiological patterns observed in the ongoing outbreak made them wonder if TMPRSS2 had something to do with the greater severity of COVID-19 symptoms observed in older people compared to children, specially if children expressed lesser levels of TMPRSS2 and ACE2. Using single-cell RNA-sequencing, which can detect the expression of genes in individual cells of tissues such as the lung, the researchers were able to track the expression of genes known to be involved in the body’s response to COVID-19 over time. Their study indicated that while the gene for ACE2 was expressed at low levels in the mouse lung, “TMPRSS2 stood out as having a really striking trajectory of increased expression during development, later using RNA in situ hybridization, using fluorescent probes, they were able to visualize expression of the TMPRSS2 gene , which increased over time in specific types of epithelial cells that line the lungs. Analyzing human lung specimen obtained across different ages of patients confirmed a similar trajectory in TMPRSS2 expression to what they’d found in mice. These findings allowed the researchers to conclusively underscore the opportunity to consider TMPRSS2 inhibition as a potential therapeutic target for SARS-CoV-2. The research was supported by several grants from National Institutes of Health and the background work for this paper was built upon the collaborative efforts of the Human Cell Atlas (HCA) Lung Biological Network, The Vanderbilt COVID-19 Consortium Cohort, a multi-disciplinary effort to understand more fully why some people are at greater risk of COVID-19 infection and illness. Story source:
Bryce A. Schuler, A. Christian Habermann, Erin J. Plosa, Chase J. Taylor, Christopher Jetter, Nicholas M. Negretti, Meghan E. Kapp, John T. Benjamin, Peter Gulleman, David S. Nichols, Lior Z. Braunstein, Alice Hackett, Michael Koval, Susan H. Guttentag, Timothy S. Blackwell, Steven A. Webber, Nicholas E. Banovich, Jonathan A. Kropski, Jennifer M. S. Sucre. Age-determined expression of priming protease TMPRSS2 and localization of SARS-CoV-2 in lung epithelium. Journal of Clinical Investigation, 2020; DOI: 10.1172/JCI140766 P.S. Content edited for style and length Serotonin the happy hormone act as a growth factor for the stem cells in the fetal human brain that determine brain size During the evolutionary journey, the size of the brain increased, especially in a particular part called the neocortex. The neocortex enables us to speak, dream and think. In the search of the causes underlying neocortex expansion, researchers at the Max Planck Institute of Molecular Cell Biology and Genetics in Dresden, together with colleagues at the University Hospital Carl Gustav Carus Dresden, identified a number of molecular players. These players typically act cell-intrinsically in the so-called basal progenitors, the stem cells in the developing neocortex with a pivotal role in its expansion. The researchers now report an additional, novel role of the happiness neurotransmitter serotonin which is known to function in the brain to mediate satisfaction, self-confidence and optimism – to act cell-extrinsically as a growth factor for basal progenitors(BPs) in the developing human, but not mouse, neocortex (Ncx). Due to this new function, placenta-derived serotonin likely contributed to the evolutionary expansion of the human neocortex. The team of Wieland Huttner at the Max Planck Institute of Molecular Cell Biology and Genetics, who is one of the institute’s founding directors, has investigated the cause of the evolutionary expansion of the human neocortex in many studies. In a new study from his lab focuses on the role of the neurotransmitter serotonin, the happiness neurotransmitter, because it transmits messages between nerve cells that contribute to well-being and happiness, for a potential role in brain development during in the developing embryo. In humans and mice the placenta produces serotonin, which then reaches the brain via the blood circulation, however, the function of this placenta-derived serotonin in the developing brain has been unknown. The findings of the study has been published in the Cell Press journal Neuron, their study revealed that the serotonin receptor HTR2A was expressed in fetal human, but not embryonic mouse, neocortex (Ncx). Serotonin needs to bind to this receptor in order to activate downstream signaling. To explore if this receptor could be one of the keys to the question of why humans have a bigger brain.” The researchers induced the production of the HTR2A receptor in embryonic mouse neocortex, and found that serotonin, by activating this receptor, caused a chain of reactions that resulted in the production of more basal progenitors in the developing brain. More basal progenitors can then increase the production of cortical neurons, which paves the way to a bigger brain. Conversely, CRISPR/Cas9-mediated knockout of endogenous HTR2A in embryonic ferret Ncx reduces BP proliferation. Pharmacological activation of endogenous HTR2A in fetal human Ncx ex vivo increases BP proliferation via HER2/ERK signaling. Hence, 5-HT emerges as an important extrinsic pro-proliferative signal for BPs, which may have contributed to evolutionary Ncx expansion. Significance for brain development and evolution According to Wieland Huttner, supervisor of the study, "the present study uncovers a novel role of serotonin as a growth factor for basal progenitors in highly developed brains, notably humans and implicates serotonin in the expansion of the neocortex during development and human evolution". He continues: “Abnormal signaling of serotonin and a disturbed expression or mutation of its receptor HTR2A have been observed in various neurodevelopmental and psychiatric disorders, such as Down syndrome, attention deficit hyperactivity disorder and autism. Our findings may help explain how malfunctions of serotonin and its receptor during fetal brain development can lead to congenital disorders and may suggest novel approaches for therapeutic avenues.” Story Source:
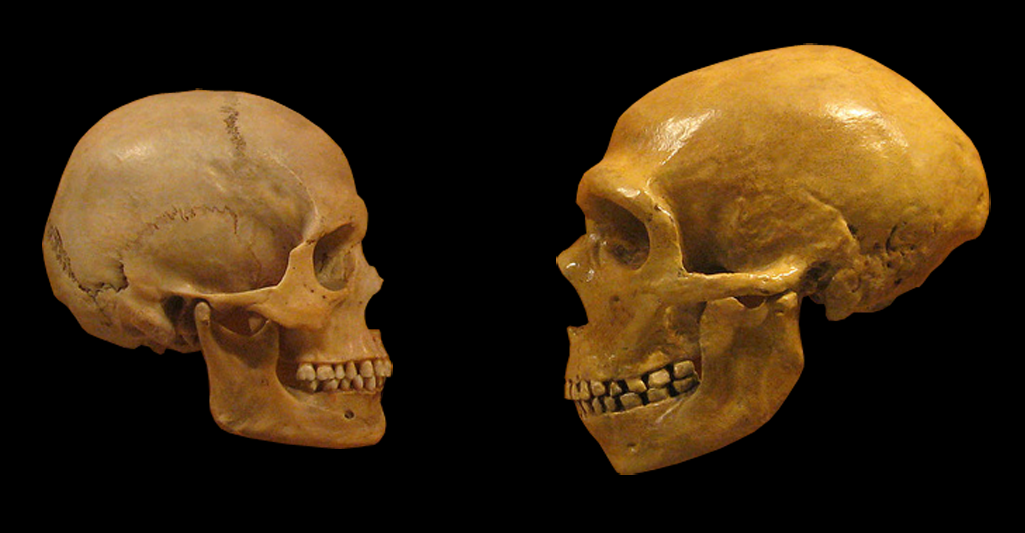
Lei Xing, Nereo Kalebic, Takashi Namba, Samir Vaid, Pauline Wimberger, Wieland B. Huttner. Serotonin Receptor 2A Activation Promotes Evolutionarily Relevant Basal Progenitor Proliferation in the Developing Neocortex. Neuron, 2020; DOI: 10.1016/j.neuron.2020.09.034 The replacement of Neanderthal by modern man is typically attributed to environmental pressure or superiority of modern humans with respect to competition for resources. According to a new study small populations, inbreeding, and random demographic fluctuations could have been enough to cause Neanderthal extinction. The study was conducted by Krist Vaesen et. al from the Eindhoven University of Technology, which is published in the open-access journal PLOS ONE. One of the biggest conundrums of palaeoanthropology is the demise of Neanderthals approximately 40000 years ago. According to a general agreement the extinction event occurred after a long period of largely separated coexistence coinciding with migration events starting around ~60 000 years ago by Anatomically Modern Humans (AMHs) from Africa into the Near East and Europe following which AMHs took over the territories previously occupied by our sister species. However, the causes of Neanderthal extinction has been attributed to a wide variety of intensely debated factors, including climatic change, epidemics, a disputed theory is of supposed "superiority" of AMHs over Neanderthals in competing for the same resources. The investigators argue that no such contested factors might be needed to account for the demise of Neanderthals. They present two independent models that capture the internal dynamics of Neanderthal populations―the models thus ignore, among other things, competitive interactions with AMHs―and that suggest that the disappearance of Neanderthals might have resided in the small size of their population(s) alone. Accordingly, the study substantiates the suggestion, that it may simply be the case that Neanderthal populations declined below their minimum viable population threshold. The study models of the investigation presented three basic factors that, according to conservation biology, would put such small populations at risk of extinction: inbreeding, Allee effects, and stochasticity. Inbreeding depression refers to the reduction in fitness of individuals that arise from matings between genetic relatives, matings thus that are more likely to occur in small populations. Inbreeding, which seems to have been common in Neanderthals might lead to a lower fitness because it increases the chances of the expression of recessive, deleterious traits and because homozygotes often have a general disadvantage relative to heterozygotes. Harris and Nielsen estimate that, due to inbreeding, Neanderthals had at least 40% lower fitness than modern humans on average. Allee effects refer to the effects that population density has on reproduction and, thus, on population growth. At lower densities, growth rates might drop due to problems in mate-finding, and to several problems that highly cooperative species, such as Neanderthals, are particularly susceptible to, including low availability of helpers in cooperative hunting, defending kills from kleptoparasites, and allo-parenting. Finally, stochastic, annual fluctuations in births, deaths, and sex ratios are more likely to place smaller populations on a trajectory towards extinction than bigger ones. The models indicate that these factors alone could have resulted in Neanderthal extinction, even if Neanderthals and AMHs were identical in terms of individual-level traits that are deemed relevant to persistence or extinction (e.g., cognitive and technological ability, sociality). The results of the study support the hypothesis that the disappearance of Neanderthals might have been the result of demographic factors alone, that is, the result merely of the internal dynamics that operate in small populations. The authors said, "The present study which provides an explanation solely in terms of the internal dynamics of the Neanderthal population, as the one presented here, serves as a null hypothesis against which competing, and less parsimonious, hypotheses are to be assessed. Regardless of whether external factors (climate or epidemics) or factors related to resource competition played a role in the actual demise of Neanderthals, our study suggests that any plausible explanation of the demise also needs to incorporate demographic factors as key variables." Thus, the study finally indicates that the arrival of AMHs would have been a contributory factor rather than the cause of the extinction. Importantly, population-level characteristics―e.g., many of the characteristics that conservation biology has shown to be critical for a species’ persistence, including population size, species distribution, intraspecific variability, and patterns of dispersal―might also account for the successful range expansion of AMHs. In other words, our species’ success need not be the result of superiority in its individual-level traits. Source:
Materials provided by PLOS One, Content edited for style and length. Journal Reference:
 Researchers examine the decline in average body temperature among healthy adults over the past two decades. For over two centuries after since German physician Carl Wunderlich established 98.6°F as the standard “normal” body temperature, it has been used by parents, doctors and other health care workers alike as the measure by which fevers — and often the severity of illness — have been assessed. however, and in more recent years, lower body temperatures have been widely reported in healthy adults. A 2017 study among 35,000 adults in the United Kingdom found average body temperature to be lower (97.9°F), and a 2019 study showed that the normal body temperature in Americans (those in Palo Alto, California, anyway) is about 97.5°F. A multinational team of physicians, anthropologists and local researchers led by Michael Gurven, UC Santa Barbara professor of anthropology and chair of the campus’s Integrative Anthropological Sciences Unit, and Thomas Kraft, a postdoctoral researcher in the same department, have found a similar decrease among the Tsimane, an indigenous population of forager-horticulturists in the Bolivian Amazon. In the 16 years since Gurven, co-director of the Tsimane Health and Life History Project, and fellow researchers have been studying the population, they have observed a rapid decline in average body temperature — 0.09°F per year, such that today Tsimane body temperatures are roughly 97.7°F. “In less than two decades we’re seeing about the same level of decline as that observed in the U.S. over approximately two centuries,” said Gurven. Their analysis is based on a large sample of 18,000 observations of almost 5,500 adults, and adjust for multiple other factors that might affect body temperature, such as ambient temperature and body mass. The investigators have published their findings in the journal Sciences Advances, which states that: “The provocative study showing declines in normal body temperature in the U.S. since the time of the Civil War was conducted in a single population and couldn’t explain why the decline happened, but it was clear that something about human physiology could have changed. One leading hypothesis is that we’ve experienced fewer infections over time due to improved hygiene, clean water, vaccinations and medical treatment. Gurven comments, "In our study, we were able to test that idea directly. We have information on clinical diagnoses and biomarkers of infection and inflammation at the time each patient was seen. While some infections were associated with higher body temperature, adjusting for these did not account for the steep decline in body temperature over time, Gurven noted. “And we used the same type of thermometer for most of the study, so it’s not due to changes in instrumentation,” he said. Added Kraft, “No matter how we did the analysis, the decline was still there. Even when we restricted analysis to the <10% of adults who were diagnosed by physicians as completely healthy, we still observed the same decline in body temperature over time.” A key question, then, is why body temperatures have declined over time both for Americans and Tsimane. Extensive data available from the team’s long-term research in Bolivia addresses some possibilities. “Declines might be due to the rise of modern health care and lower rates of lingering mild infections now as compared to the past,” Gurven explained. “But while health has generally improved over the past two decades, infections are still widespread in rural Bolivia. Our results suggest that reduced infection alone can’t explain the observed body temperature declines.” It could be that people are in better condition, so their bodies might be working less to fight infection, he continued. Or greater access to antibiotics and other treatments means the duration of infection is shorter now than in the past. Consistent with that argument, Gurven said, “We found that having a respiratory infection in the early period of the study led to having a higher body temperature than having the same respiratory infection more recently.” It’s also possible that greater use of anti-inflammatory drugs like ibuprofen may reduce inflammation, though the researchers found that the temporal decline in body temperature remained even after their analyses accounted for biomarkers of inflammation. “Another possibility is that our bodies don’t have to work as hard to regulate internal temperature because of air conditioning in the summer and heating in the winter,” Kraft said. “While Tsimane body temperatures do change with time of year and weather patterns, the Tsimane still do not use any advanced technology for helping to regulate their body temperature. They do, however, have more access to clothes and blankets.” The researchers were initially surprised to find no single “magic bullet” that could explain the decline in body temperature. “It’s likely a combination of factors — all pointing to improved conditions,” Gurven said. According to Gurven, the finding of lower-than-expected body temperatures in the U.S., and the decline over time, had a lot of people scratching their heads. Was it a fluke? In this study, Gurven and his team confirm that body temperatures below 98.6°F are found in places outside the U.S. and the U.K. “The area of Bolivia where the Tsimane live is rural and tropical with minimal public health infrastructure,” he noted. “Our study also gives the first indication that body temperatures have declined even in this tropical environment, where infections still account for much morbidity and mortality.” As a vital sign, temperature is an indicator of what’s occurring physiologically in the body, much like a metabolic thermostat. “One thing we’ve known for a while is that there is no universal ‘normal’ body temperature for everyone at all times, so I doubt our findings will affect how clinicians use body temperature readings in practice” said Gurven. Despite the fixation on 98.6°F, most clinicians recognize that ‘normal’ temperatures have a range. Throughout the day, body temperature can vary by as much as 1°F, from its lowest in the early morning, to its highest in the late afternoon. It also varies across the menstrual cycle and following physical activity and tends to decrease as we age. But by linking improvements in the broader epidemiological and socioeconomic landscape to changes in body temperature, the study suggests that information on body temperature might provide clues to a population’s overall health, as do other common indicators such as life expectancy. “Body temperature is simple to measure, and so could easily be added to routine large-scale surveys that monitor population health,” Gurven said. Story Source:
University of California - Santa Barbara. Original article written by Andrea Estrada. Note: Content edited for style and length. Journal Reference:
 Our brains are made up of billions of incredibly diverse neurons. They first arise in the developing brain when stem cells stop self-renewing and differentiate into a particular type of neuron. This process, called neurogenesis, is precisely regulated to give rise to the enormous complex structure that is our brain. It is thought that small differences in the way neural stem cells generate neurons are at the origin of the dramatic increase in the size and complexity of our brain. To gain insight in this complex process, prof. Pierre Vanderhaeghen (VIB-KU Leuven, ULB) and his colleagues examined the mitochondria, small organelles that provide energy in every cell in the body, including the developing brain. “Diseases caused by defects in mitochondria lead to developmental problems in many organs, in particular the brain,” explains Vanderhaeghen, a specialist in stem cell and developmental neurobiology. “We used to think that this was related to the crucial function of mitochondria to provide energy to the cells, but this is only part of the story: recent work in stem cells suggests that mitochondria have a direct influence on organ development. We have tested whether and how this could be the case in the brain.” Fission and fusion Together with his team, he explored whether and how mitochondrial remodeling is coupled with neuronal fate commitment during neurogenesis. “Mitochondria are highly dynamic organelles, that can join together (fusion) or split up (fission), and we know these dynamics are associated with fate changes in various types of stem cells,” says Vanderhaeghen. Ryohei Iwata, a postdoctoral researcher in the Vanderhaeghen lab, developed a new method to watch mitochondria in great detail as the neural stem cells are ‘caught in the act’ to become neurons. “We found that shortly after stem cells divide, the mitochondria in daughter cells destined to self-renew will fuse, while those in daughter cells that become neurons show high levels of fission instead,” says Ryohei Iwata. But this was not just a coincidence: indeed, the researchers could show that increased mitochondrial fission in fact promotes differentiation to a neuronal fate, while mitochondrial fusion after mitosis redirects daughter cells towards self-renewal. Time window So mitochondrial dynamics are important to become a neuron—but there is more. “We found that the influence of mitochondrial dynamics on cell fate choice is limited to a very specific time window, right after cell division,” says Pierre Casimir, a PhD student in Vanderhaeghen’s lab. “Interestingly, the restricted time window is twice as long in humans compared to mice.” “Previous findings were primarily focused on fate decision of neural stem cells before they divide, but our data reveal that cell fate can be influenced for a much longer period, even after neural stem cell division,” says Vanderhaeghen. This may have interesting implications in the emerging field of cell reprogramming, where scientists try to convert non-neuronal cells directly in neuronal cells for therapeutic purposes for instance. “Since this period of plasticity is much longer in human cells compared to mouse cells, it is tempting to speculate that it contributes to the increased self-renewal capacity of human progenitor cells, and thus to the uniquely developed brain and cognitive abilities of our species. It is fascinating to think that mitochondria, small organelles that have evolved in cells more than a billion years ago, might have contributed to the recent evolution of the human brain.” Story Source:
News article from The Flanders Institute for Biotechnology. Note: Content edited for style. Journal Reference: Ryohei Iwata, Pierre Casimir, Pierre Vanderhaeghen. Mitochondrial dynamics in postmitotic cells regulate neurogenesis. Science, 2020 DOI: 10.1126/science.aba9760  Pluripotent stem cells have the ability to self-renew and give rise to all other types of cells and organs in the body. The developmental patterns and characteristics are controlled by a network of regulatory genes and molecules, but little is known about how this network has evolved across mammals. The recent study by Kyoto University researchers across 48 mammalian species published in the journal Genome Biology and Evolution. The study group observed that the genes regulating pluripotent stem cells in mammals are surprisingly similar. In the study conducted by Ken-ichiro Kamei of Kyoto University’s Institute for Integrated Cell-Material Sciences (iCeMS), with Miho Murayama and Yoshinori Endo of the Wildlife Research Center, compared 134 gene sets belonging to the pluripotency gene regulatory networks of 48 mammalian species. They found that this network has been highly conserved across species, meaning genetic sequences have remained relatively unchanged over the course of evolution. This high degree of conservation explains why human genetic sequences can reprogram other mammalian tissue cells to turn into pluripotent stem cells. However, since it is also evident that the regulating networks differ across mammals, there might be more efficient combinations of reprogramming factors for each species. Improving techniques for deriving induced pluripotent stem (iPS) cells from mammalian cells, including those from endangered species, could provide a big boost to research and conservation. “We have been trying to generate induced pluripotent stem cells from various mammalian species, such as the endangered Grévy’s zebra and the bottlenose dolphin,” says Kamei. Interestingly, the team found relatively high evolutionary changes in genes just downstream of one of the core gene regulatory networks. “This could indicate that mammalian pluripotent stem cells have diversified more than we thought,” says Inoue-Murayama. The differences between gene regulatory networks in mammalian pluripotent stem cells might also be associated with unique adaptions. For example, the naked mole rat has been positively selected for a pluripotency regulatory gene that could be involved in giving it its extraordinary longevity and cancer resistance. The gene might also be involved in the development of the extremely sensitive hairs that help them navigate underground. The researchers also found evidence of positive selection for certain pluripotency gene regulatory network genes involved in the adaptation of large animals, such as the minke whale, the African elephant and the flying fox, to their environments. Surprisingly, these same genes are associated with cancer in other mammals. Since these large animals are known for being relatively resistant to cancer, the researchers suggest that the adaptive alterations these genes underwent in these animals somehow also changed some of their functions, thus giving this group a degree of cancer resistance. The researchers say the study is among the first to compare the pluripotency gene regulatory networks across major taxa, and could be applicable to evolutional biology studies and for facilitating and improving the generation of induced pluripotent stem cells from new species. Story Source:
Kyoto University. Content edited for style and length Journal Reference: Yoshinori Endo, Ken-ichiro Kamei, Miho Inoue-Murayama. Genetic signatures of evolution of the pluripotency gene regulating network across mammals. Genome Biology and Evolution, 2020; DOI: 10.1093/gbe/evaa169  Recent findings by researchers at Yale points towards a host of genetic risk factors that explains susceptibility to the debilitating symptoms of post-traumatic stress disorder (PTSD) in veterans. The Yale-led study published on Sept. 30 in the journal Biological Psychiatry has now identified a social factor that can mitigate these genetic risks: the ability to form loving and trusting relationships with others. The study is one of the first to explore the role of nurture as well as nature in its investigation of the biological basis of PTSD. “We exist in a context. We are more than our genes,” said Yale’s Robert H. Pietrzak, associate professor of psychiatry and public health, and senior author of the study. Pietrzak is also director of the Translational Psychiatric Epidemiology Laboratory of the U.S. Department of Veterans Affairs National Center for PTSD. Like many genetic studies on mental disorders such as depression, anxiety, and schizophrenia, PTSD studies have revealed numerous genetic risk factors that contribute to the severity of the disorder. For instance, a previous study of more than 165,000 U.S. military veterans led by Yale’s Joel Gelernter, the Foundations Fund Professor of Psychiatry and professor of genetics and of neuroscience, found variants in eight separate regions of the genome that help predict who is most likely to experience the repeated disturbing memories and flashbacks that are hallmark symptoms of PTSD. In the new study, Pietrzak, Gelernter, and colleagues looked at psychological as well as genetic data collected from the National Health and Resilience in Veterans Study, a national sample of U.S. military veterans supported by the National Center for PTSD. The researchers specifically focused on a measure of attachment style — the ability or inability to form meaningful relations with others — as a potential moderator of genetic risk for PTSD symptoms. Individuals with a secure attachment style perceive relationships as stable, feel that they are worthy of love and trust, and are able to solicit help from others. Those with an insecure attachment style report an aversion to or anxiety about intimacy with others, and have difficulty asking for help from others. They found that the ability to form secure attachments essentially neutralized the collective effects of genetic risk for PTSD symptoms. The impact was particularly pronounced in a variant of the IGSF11 gene, which has been linked to synaptic plasticity or the ability of the brain to form new connections between brain cells. Pietrzak noted that deficits in synaptic plasticity have also been linked to PTSD, depression, and anxiety, among other mental disorders. The findings illustrate the importance of integrating environmental and social as well as genetic factors in the study of PTSD and other mental disorders, the authors said. “Social environmental factors are critical to informing risk for PTSD and should be considered as potential moderators of genetic effects,” he said. “The ability to form secure attachments is one of the strongest protective factors for PTSD and related disorders.” The attachment styles may moderate polygenic risk for PTSD symptoms, along with the effects of a novel locus implicated in synaptic transmission and plasticity which may serve as a possible biological mediator of this association. These findings may help inform interpersonally-oriented treatments for PTSD for individuals with high polygenic risk for this disorder and will help predict who is at greater risk of experiencing severe symptoms of PTSD, the study also suggest that psychological treatments targeting interpersonal relationships may help mitigate PTSD symptoms in veterans with elevated genetic risk for this disorder . Amanda Tamman, formerly of Yale and now a Ph.D. student in clinical psychology at St. John’s University, is first author of the paper. Story Source:
Yale News bulletin written by By Bill Hathaway. Article modified for style and clarity, Image source: Wikimedia Commons Journal Article: Amanda J.F. Tamman, Frank R. Wendt, Gita A. Pathak, John H. Krystal, Janitza L. Montalvo-Ortiz, Steven M. Southwick, Lauren M. Sippel, Joel Gelernter, Renato Polimanti, Robert H. Pietrzak. Attachment style moderates polygenic risk for posttraumatic stress in United States military veterans: Results from the National Health and Resilience in Veterans Study. Biological Psychiatry, 2020; DOI: 10.1016/j.biopsych.2020.09.018 In biology all of us share a basic question in biology, what properties are shared among organisms? Comparative genomics and genome sequencing allows comparison of organisms at DNA and protein levels, and sequence alignment is a way of arranging the sequences of DNA, RNA, or protein to identify regions of similarity that may be a consequence of functional, structural, or evolutionary relationships between the sequences. The sequence Comparisons can be used to: - Find evolutionary relationships between organisms - Identify functionally conserved sequences - Identify corresponding genes in human and model - organisms: develop models for human diseases Thus, sequence alignment is an important first step toward structural and functional analysis of newly determined sequences to draw functional and evolutionary inference. The sequence alignment is made between a known sequence and unknown sequence or between two unknown sequences. The known sequence is called reference sequence, and the unknown sequence is called query sequence. To proceed with the alignment process the sequences are either aligned in group of two which is called pair-wise alignment) or more than two known as, multiple sequence alignment) sequences by searching for a series of individual characters or character patterns that are in the same order in the sequences. Identical or similar characters are placed in the same column, and non-identical characters can either be placed in the same column as a mismatch or opposite a gap in the other sequence. In an optimal alignment, non-identical characters and gaps are placed to bring as many identical or similar characters as possible into vertical register. Depending upon the region of comparison, alignments are divided into two types of viz. global and local. Global Alignment Global alignment program is based on Needleman-Wunsch algorithm In global alignment, two sequences to be aligned are assumed to be generally similar over their entire length. Alignment is carried out from beginning to end of both sequences to find the best possible alignment across the entire length between the two sequences. The two sequences are treated as potentially equivalent. Goal for Global alignment: Identify conserved regions and differences, and it is applied for either comparing two genes with same function. or for comparing two sequences for conserved regions. Local Alignment Local alignment program are based on Smith-Waterman, algorithm. Local alignment does not assume that the two sequences in question have similarity over the entire length, rather, it only finds local regions with the highest level of similarity between the two sequences and aligns these regions without regard for the alignment of the rest of the sequence regions. There are three primary methods of producing local alignments, dot-matrix methods, dynamic programming, and word or k-tuple method. Goal for local alignment: The goal for local alignment is to check whether a substring in one sequence aligns well with a substring in the other, and it is applied for searching local regions of similarities in large sequences (e.g., newly sequenced genomes). or for searching conserved domains or motifs. Significance of sequence alignment Sequence alignment is useful for discovering functional, structural, and evolutionary information in biological sequences. However, it is important to obtain the best possible or “optimal” alignment to discover this information. Sequences that are very much alike, or “similar” in the parlance of sequence analysis, probably have the same function or there may have been a common ancestor sequence, and thus, the sequences are then defined as being homologous. The alignment indicates the changes that could have occurred between the two homologous sequences during the course of evolution. Now let us learn more about sequence alignment using this video tutorial. BLAST (basic local alignment search tool) is an algorithm and program for comparing primary biological sequence information, such as the amino-acid sequences of proteins or the nucleotides of DNA or RNA sequences. BLAST performs “local” alignments, and this is particularly helpful when working with one or more functional domains occurring within a protein. The BLAST algorithm is tuned to find these domains or shorter stretches of sequence similarity. Moreover, the local alignment approach also means that an mRNA can be aligned with a piece of genomic DNA, as its is frequently required in genome assembly and analysis. BLAST works by finding regions of local similarity between sequences comparing nucleotide or protein sequences to sequence databases and it also calculates the statistical significance of matches, and displays a “expect value” or e-value that estimates how many matches would have occurred at a given score by chance, which can aid a user in judging how much confidence to have in an alignment. Uses of BLAST BLAST can be used for several purposes such as identification of species, locating domains, establishing phylogeny, DNA mapping, and Sequence comparisons. Identification of species: With the use of BLAST, we can correctly identify a species or find homologous species. This can be useful, for example, when you are working with a DNA sequence from an unknown species. Locating domains: When working with a protein sequence you can input it into BLAST, to locate known domains within the sequence of interest. Establishing phylogeny: Using the results received through BLAST you can create a phylogenetic tree using the BLAST web-page. Phylogenies based on BLAST alone are less reliable than other purpose-built computational phylogenetic methods, so should only be relied upon for "first pass" phylogenetic analyses. DNA mapping: When working with a known species, and looking to sequence a gene at an unknown location, BLAST can compare the chromosomal position of the sequence of interest, to relevant sequences in the database(s). NCBI has a "Magic-BLAST" tool built around BLAST for this purpose.[30] Sequence Comparison: When working with genes, BLAST can locate common genes in two related species, and can be used to map annotations from one organism to another. Therefore BLAST has proven itself to be an important tool for studying functional and evolutionary relationships between sequences as well as help identify members of gene families. One of the aim of biologiks is to make learning concepts of biology interesting and working on that line it brings me immense pleasure to share few of the educational resources that I have created for my student friends in the form of YouTube videos at our official YouTube channel. Do check them out, share with your friends and leave your comments, if you find it helpful, as you, the students and learners, are always my inspiration to bring up better content every time.
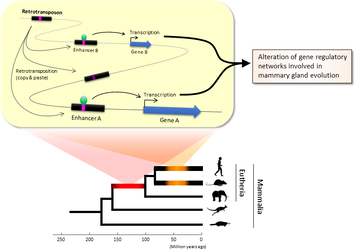 Figure 1. Retrotransposons have been co-opted to act as enhancers in the mammalian genome. Transposable elements known as retrotransposons duplicate themselves via a copy-and-paste mechanism. Tens of thousands of these sequences were found to contain binding sites for proteins that act as master regulators of mammary gland development. Figure 1. Retrotransposons have been co-opted to act as enhancers in the mammalian genome. Transposable elements known as retrotransposons duplicate themselves via a copy-and-paste mechanism. Tens of thousands of these sequences were found to contain binding sites for proteins that act as master regulators of mammary gland development. Transposable elements (TEs), so-called selfish DNA sequences, are known to be capable of moving around the genome through cut-and-paste or copy-and-paste mechanisms, and our human genome contains approx 4.5 million copies of these TEs. This can not be termed as one obscure event as they account for 30-50% of mammalian DNA. The trnasposable elements have been traditionally considered as genetic freeloaders hitchhiking along in the genome without providing any benefit to the host organism. More recently, however, scientists have begun to uncover cases in which TE sequences have been co-opted by the host to provide a useful function, such as encoding part of a host protein. In a recent study published in the journal Nucleic Acids Research, Professor Hidenori Nishihara from Department of Life Science and Technology, Tokyo Institute of Technology, who has undertaken one of the most comprehensive analyses of TE sequence co-option to date, uncovers tens of thousands of potentially co-opted TE sequences and the findings suggest that the TEs might have played a key role in mammalian evolution. Talking about his research Professor Nishihara says that "I was specifically interested in the potential influence of TE sequences on the evolution of the mammary gland, an organ that is responsible for producing milk and is, as the name suggests, a key distinguishing feature of mammals." To identify potentially co-opted TE sequences, Dr. Nishihara used four proteins—ERα, FoxA1, GATA3, and AP2γ—that bind to DNA to regulate the production of proteins involved in mammary gland development, and located all of the DNA sequences in the genome to which these proteins bind. Surprisingly, 20–30% of all of the binding sites across the genome were located in TEs, with as many as 38,500 TEs containing at least one binding site. The majority of these were in a copy-and-paste type of TE known as a retrotransposon, which duplicates itself, leaving a new copy in a new location. The TE-derived binding site sequences were more conserved across species than expected, indicating that they are being preserved by evolution because they serve some important function. Dr. Nishihara believes that these TE sequences have been co-opted to serve as enhancers, DNA elements that increase the transcription of nearby genes (Fig. 1). By binding to one of the four master regulators of mammary gland development, these enhancers ultimately increase the production of proteins involved in mammary gland development. Dr. Nishihara then investigated when in mammalian evolution these TE sequences were acquired and found two distinct phases of acquisition: roughly 60–70% were acquired in the ancestor of all placental mammals (Eutheria), while 10–20% could be traced back to the ancestor of New World monkeys (Simiiformes) (Fig. 2, left). In addition, there appeared to be another wave of acquisition of ERα binding sites in the ancestor of mice and rats (Muridae) (Fig. 2, right). Thus, by providing a vast number of potential regulatory element binding sites throughout the genome, TEs may have had a substantial impact on the emergence of the mammary gland and its evolution within mammals. 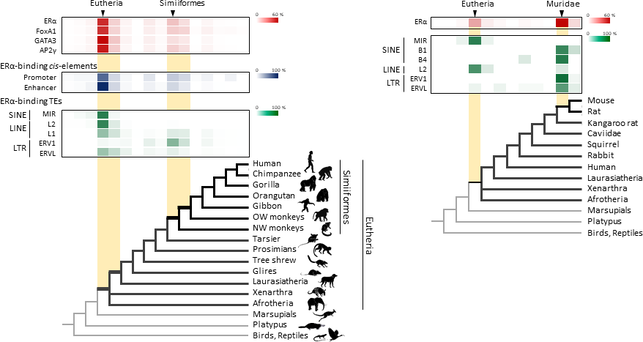 Figure 2. Transposable element-derived binding sites were acquired during distinct phases in mammalian evolution. Left: Among the TE-derived binding sites identified, 60–70% were acquired in the ancestor of placental mammals (Eutheria), while 10–20% were acquired in the ancestor of New World monkeys (Simiiformes). Right: Many ERα binding sites were also acquired in the ancestor of mice and rats (Muridae). Dr. Nishihara's study sheds light on the deep involvement of TEs in the evolution of mammary gland regulatory elements. However, it remains unclear how common this mode of TE-mediated regulatory network evolution is. Dr. Nishihara, at least, believes that the mammary gland is not unique in this respect. He notes that, "in addition to mammary glands, mammals share many features, such as the neocortex, closed secondary palate, and hair. I expect future research to uncover many additional kinds of TEs that have been similarly involved in the evolution of these features in mammals." References
One of the oldest desire of man kind is to increase its life expectancy and if possible be immortal. In pursuit of this dream countless have spent their lives searching for elixir of immortality to fountains of youth, often leading to pain and animosity between fellow humans. Japan, a country owing to its cultural, behavioral and numerous genetic factors has been blessed with many centenarians and currently has the greatest number of known centenarians of any nation with 67,824 according to their 2017 census, along with the highest proportion of centenarians at 34.85 per 100,000 people. Thus Japan becomes the primal choice for conducting studies reflecting on the secrets of longer life and it can be performed with accuracy and a larger statistical sample size compared to other nations. In a latest set of findings research teams of scientists from the RIKEN Center for Integrative Medical Science (IMS) and Keio University School of Medicine in Japan have shown us that all the while we have been looking in the wrong direction and the solution and clue to long life was within our body's own defense / the immune system. Supercentenarians -- meaning people over the age of 110 --as their study interest the teams have discovered an interesting finding that the supercentenarians proved to be an unique group of people having a higher count of specific immune cells cytotoxic CD4+ T-cells, when they compared their cell count with a group of supercentenarians and younger controls. They acquired a total of 41,208 cells from seven supercentenarians (an average of 5,887 per subject) and 19,994 cells for controls (an average of 3,999 per subject) from five controls aged in their fifties to eighties. The study revealed two interesting findings:
Kosuke Hashimoto of IMS, the first author of the paper, expressed the team's stand as "We were especially interested in studying this group of people, because we consider them to be a good model of healthy aging, and this is important in societies like Japan where aging is proceeding rapidly." IMS Deputy Director Piero Carninci, one of the leaders of the groups, says, "This research shows how single-cell transcription analysis can help us to understand how individuals are more or less susceptible to diseases. CD4-positive cells generally work by generating cytokines, while CD8-positive cells are cytotoxic, and it may be that the combination of these two features allows these individuals to be especially healthy. We believe that this type of cells, which are relatively uncommon in most individuals, even young, are useful for fighting against established tumors, and could be important for immunosurveillance. This is exciting as it has given us new insights into how people who live very long lives are able to protect themselves from conditions such as infections and cancer." Their research, is published in journal of Proceedings of the National Academy of Sciences (PNAS), and the study was performed by a collaboration including scientists from the RIKEN Center for Integrative Medical Sciences and Keio University School of Medicine. References
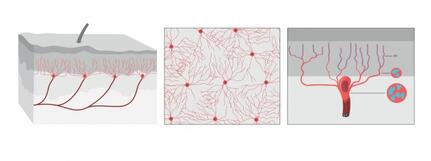 The illustration shows what the organ looks like from the side, from above the skin. Illustrator: Mattias Karlén. Reprinted with permission from Abdo et al, Science, Vol. 365, Issue 6454, pp. 695-699 (2019). The illustration shows what the organ looks like from the side, from above the skin. Illustrator: Mattias Karlén. Reprinted with permission from Abdo et al, Science, Vol. 365, Issue 6454, pp. 695-699 (2019).
Pain is one of those feelings which if not all, most of us would never like to encounter and forget our previous encounters as well. However pain has been a necessary evil as it enables our bodies to recognize the occurrence of injuries or other problems which may not be always visible to our eyes, thus preventing further damage.
The International Association for the Study of Pain's widely used definition defines pain as "an unpleasant sensory and emotional experience associated with actual or potential tissue damage, or described in terms of such damage". Before discovery of neurons responsible for pain by Charles Scott Sherrington in 1906 and the role of nociceptors, various theories were proposed to explain the origin of pain. Ancient Greeks including Hippocrates believed that it was due to an imbalance in vital fluids. Now a team of researchers from Karolinska Institutet have now discovered a new sensory receptor organ in the skin that is sensitive to hazardous environmental irritation. Conventionally pain has been thought to be initiated by activation of free nerve endings without end organs in the skin. In contrast to this paradigm, Abdo et al. discovered a previously unknown meshlike organ covering the skin that senses dangerous environmental stimuli. This organ is built from specialized glial cells with multiple long protrusions and which collectively go to make up a mesh-like organ within the epidermal-dermal border of skin. This organ is sensitive to painful mechanical damage such as pricks and pressure. The present study describes what the new pain-sensitive organ looks like, how it is organised together with pain-sensitive nerves in the skin and how activation of the organ results in electrical impulses in the nervous system that result in reflex reactions and an experience of pain. In their experiments, the researchers also blocked the organ and saw a resultant decreased ability to feel mechanical pain. "Our study shows that sensitivity to pain does not occur only in the skin's nerve fibres, but also in this recently-discovered pain-sensitive organ. The discovery changes our understanding of the cellular mechanisms of physical sensation and it may be of significance in the understanding of chronic pain," says Prof. Patrik Ernfors, professor at Karolinska Institutet's Department of Medical Biochemistry and Biophysics and chief investigator for the study. The research was carried out with financial assistance from ERC, the Swedish Research Council, the Knut and Alice Wallenberg Foundation and Welcome Trust.
References
1. Abdo H, Calvo-Enrique L, Martinez Lopez J, Song J, Zhang MD, Usoskin D, El Manira A, Adameyko I, Hjerling-Leffler J, Ernfors P. Specialized cutaneous Schwann cells initiate pain sensation. Science, 2019 DOI: 10.1126/science.aax6452 2. Image source: Karolinska Instituet 3. Source article: Karolinska Instituet 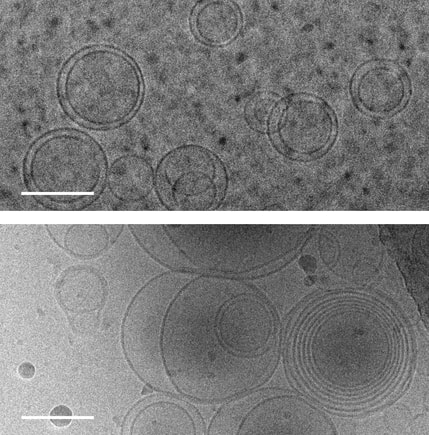 Images of membranes (circles) taken using transmission electron cryomicroscopy. Top: membranes in a solution that contains no amino acids. Bottom: membranes in a solution containing serine, an amino acid, which triggers membranes to form multiple layers of concentric membranes. Scale bars: 100 nanometers. Images of membranes (circles) taken using transmission electron cryomicroscopy. Top: membranes in a solution that contains no amino acids. Bottom: membranes in a solution containing serine, an amino acid, which triggers membranes to form multiple layers of concentric membranes. Scale bars: 100 nanometers.
The Blue planet of ours is still the only place in universe where we know life exists! However life's journey in this planet was no less than an adventurous journey when it started 4 billion years ago when the first cells formed within a primordial soup of complex, carbon-rich chemical compounds. The primitive life forms or coacervates faced a molecular conundrum as they needed charged ions to perform their basic functions however these ions could de-stabilize and disrupt the simple membranes that encapsulated the cells.
This puzzle has been solved by a team of researchers at University of Washington and their findings were published Aug. 12 in the Proceedings of the National Academy of Sciences. The team solved this puzzle using only molecules that would have been present on the early Earth. Using cell-sized, fluid-filled compartments surrounded by membranes made of fatty acid molecules, the team discovered that amino acids, the building blocks of proteins, can stabilize membranes against magnesium ions. Their results set the stage for the first cells to encode their genetic information in RNA, a molecule related to DNA that requires magnesium for its production, while maintaining the stability of the membrane. Beside explaining the mechanism how amino acid could stabilize the membranes in unfavorable condition they went beyond and demonstrated how the individual building blocks of cellular structures — membranes, proteins and RNA — could have co-localized within watery environments on the ancient Earth. “Cells are made up of very different types of structures with totally different types of building blocks, and it has never been clear why they would come together in a functional way,” said co-corresponding author Roy Black, a UW affiliate professor of chemistry and bioengineering. “The assumption was just that — somehow — they did come together.” Prof. Black teamed up with Sarah Keller, a UW professor of chemistry and an expert on membranes. Black had been inspired by the observation that fatty acid molecules can self-assemble to form membranes, and hypothesized that these membranes could act as a favorable surface to assemble the building blocks of RNA and proteins.“You can imagine different types of molecules moving within the primordial soup as fuzzy tennis balls and hard squash balls bouncing around in a big box that is being shaken,” said Keller, who is also co-corresponding author on the paper. “If you line one surface inside the box with Velcro, then only the tennis balls will stick to that surface, and they will end up close together. Roy had the insight that local concentrations of molecules could be enhanced by a similar mechanism.” The team previously had shown that the building blocks of RNA preferentially attach to fatty acid membranes and, surprisingly, also stabilize the fragile membranes against detrimental effects of salt, a common compound on Earth past and present. The team hypothesized that amino acids might also stabilize membranes. Using variety of experimental techniques — including light microscopy, electron microscopy and spectroscopy — to test how 10 different amino acids interacted with membranes. Their experiments revealed that certain amino acids bind to membranes and stabilize them. Some amino acids even triggered large structural changes in membranes, such as forming concentric spheres of membranes — much like layers of an onion. “Amino acids were not just protecting vesicles from disruption by magnesium ions, but they also created multilayered vesicles — like nested membranes,” said lead author Caitlin Cornell, a UW doctoral student in the Department of Chemistry. The researchers also discovered that amino acids stabilized membranes through changes in concentration. Some scientists have hypothesized that the first cells may have formed within shallow basins that went through cycles of high and low concentrations of amino acids as water evaporated and as new water washed in. The new findings that amino acids protect membranes — as well as prior results showing that RNA building blocks can play a similar role — indicate that membranes may have been a site for these precursor molecules to co-localize, providing a potential mechanism to explain what brought together the ingredients for life. Keller, Black and their team will turn their attention next to how co-localized building blocks did something even more remarkable: They bound to each other to form functional machines. "That is the next step,” said Prof. Black. The study was co-authored by Gary Drobny, UW professor of chemistry; Kelly Lee, UW associate professor of medicinal chemistry; UW postdoctoral researchers Mengjun Xue and Helen Litz in the Department of Chemistry, and James Williams in the Department of Medicinal Chemistry; UW graduate students Zachary Cohen in the Department of Chemistry and Alexander Mileant in the Biological Structure, Physics and Design Graduate Program; and UW undergraduate alumni Andrew Ramsay and Moshe Gordon. The research was funded by NASA, the National Institutes of Health and the National Science Foundation.
References
1. Original article byJames Urton UW news. Note: Content may be edited for style and length. 2. Image resource: Alex Mileant/Caitlin Cornell, University of Washington. |
AuthorHello! My name is Arunabha Banerjee, and I am the mind behind Biologiks. Leaning new things and teaching biology are my hobbies and passion, it is a continuous journey, and I welcome you all to join with me Archives
June 2024
Categories
All
|







 RSS Feed
RSS Feed



